At the end of the day, microbes are basically chemical factories. Through their ability to break down molecules, they were able to terraform the face of the Earth. Plants are relying on complex microbial communities to metabolize soil nutrients, and almost all animal life on Earth has their own personal chemical bioreactor to process nutrients and supply their energy - we call this the gut microbiome.
The specific chemical reactions that microbes mediate in their natural environment are complex and are mostly hidden from us. The vast majority of microbes have not been cultured, limiting our ability to poke and probe these microbes in a laboratory setting. In addition, laboratory conditions are often not representative of the chemical processes occurring in the natural environments. There are estimated to be millions of small molecules, of which we can only identify a few thousand of them. Fortunately, due to rapid advances in sequencing and mass spectrometry, we no longer need to grow microbes in a lab to observe them. Instead, we can count microbes and metabolites within an environment and attempt to infer how they change across experimental conditions to enable more focused experiments.
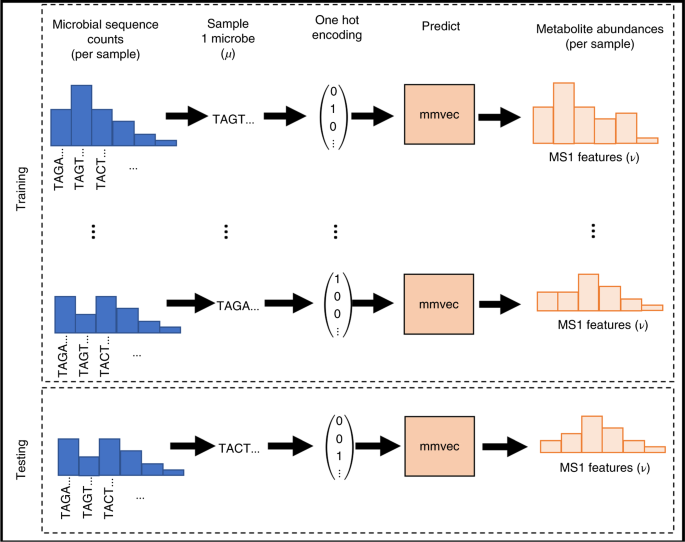
However, designing statistical tools to analyze these datasets is not trivial - we showed in our paper the vast majority of tools available to learn microbe-metabolite interactions perform comparable to random chance. To address this problem, we showed that analogy reasoning methods used in natural language processing can be adapted to infer relationships between microbes and metabolites. Our tool mmvec (short for microbe-metabolite vectors) was developed to learn a coordinate system to pinpoint the relationships between microbes and metabolites based on how often they are found within the same environment.

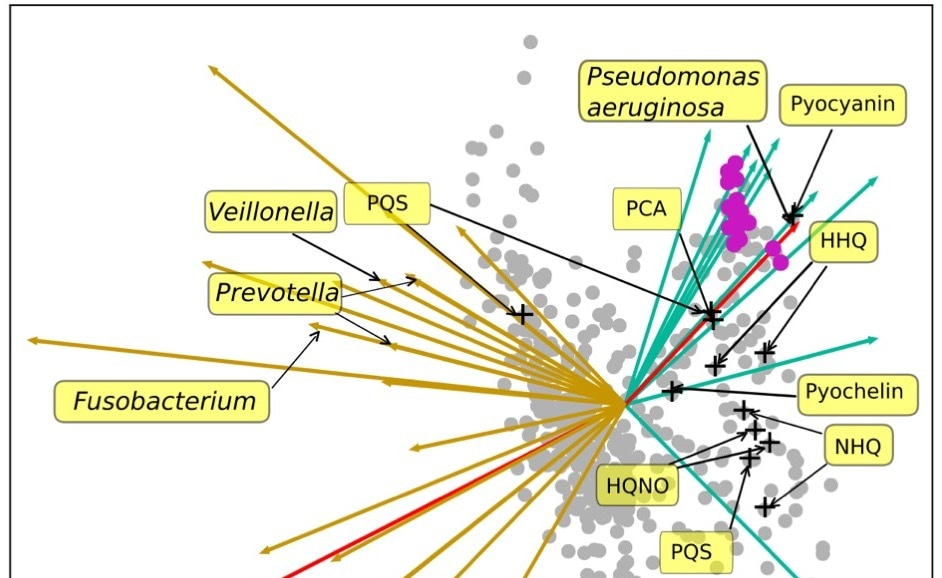
Figure 1: Above is a 2D representation of microbes and metabolites from samples collected from cystic fibrosis patients. Points represent molecules and arrows represent microbes. The closer the microbes / metabolites are together, the more likely they will be found in the same environment together.
Our tool can be found at https://github.com/biocore/mmvec
Furthermore, our tool is available through qiime2, so don’t hesitate to post on the qiime2 forums with the mmvec tag if you have any questions.






Please sign in or register for FREE
If you are a registered user on Research Communities by Springer Nature, please sign in